Behind the scenes: how do soil bacterial communities assemble?
Senka Causevic and Janko Tackmann
How do microbial soil ecosystems assemble? This question brought together two very different research groups from the NCCR Microbiomes community, leading to work that was finally published in March 2022(1). Here, we present the story behind this publication.
Soils are vast ecosystems, teeming with life and full of microbial diversity. Dense networks of microbes ensure soil health and functioning, with impacts on soil fertility, global biogeochemical cycles, and more (2). Previous research indicated that microbial communities in different environments remain distinct, creating “signature” communities (also called microbiomes, such as ocean microbiomes, human gut microbiomes, etc.) (3). Further, in the case of soil, the most abundant taxa are generally shared among soil microbiomes sampled across the globe (4,5). This led us to wonder: how do soil communities form such consistent signatures? Understanding how microbiomes assemble and shape their environment could allow us to make them more resilient or even restore them after drought (6), pollution (7) and other stresses, which could be helpful in these times of major global changes.
To tackle this question, we joined forces within the NCCR Microbiomes community, combining the soil and synthetic community expertise of the van der Meer group with the computational know-how of the von Mering group.
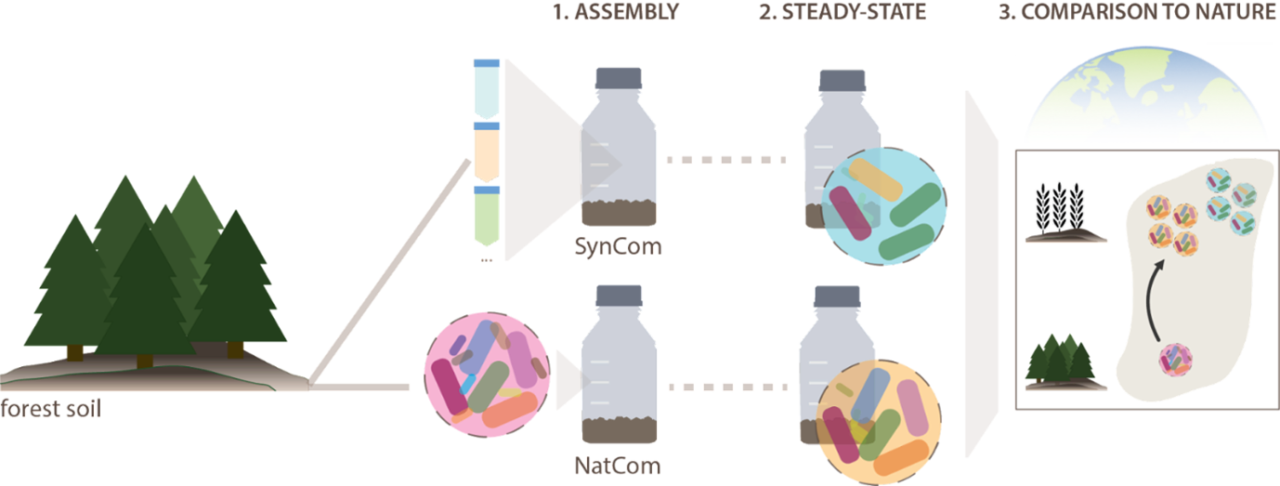
As a first step, in order to strip away natural complexity and mechanistically approach our question, we turned towards synthetic communities (Figure 1). To this end, we leveraged two complementary design approaches, each with its own advantage. In a bottom-up approach (SynCom), we constructed experimentally tractable communities by combining 21 representative soil isolates, while in the top-down approach (NatCom), we derived more diverse and representative, but less controllable communities directly from natural forest soil by detaching and purifying the cells from soil particles. In addition, we designed a culture system to grow microbes in soil. Our method of soil culturing provides: i) a fragmented micro-habitat structure and ii) the complex nutrient profile of natural soils. Both factors, we were thinking, would influence interspecific interactions, would create ecological niches, might favor diversity, and might instigate emergent community behaviors. These characteristics are not captured by the conventional method of culturing in liquid suspensions. We then inoculated SynCom and NatCom communities into the soil systems in a large number of replicates and investigated the development of the communities and their composition over time, and also across multiple passages of soil growth and mixing small batches of soil-grown community with fresh (sterile) soil (see Figure 2).
A Case Study in Collaboration
When Senka and Jan wanted to compare their results from experimental soils to “the big picture,” they turned to Janko and Christian, and the global collection of samples amassed by the Microbe Atlas Project.
They attribute their successful collaboration to:
Alternating moments of discovery
- Microbe Atlas was designed to enable big-picture comparisons, and Senka and Jan’s request for soil-specific analysis was a model test case.
- Senka and Jan were surprised by how their soil community fit into the global picture.
- Janko was pleased to see that Microbe Atlas could be a tool for generating hypotheses and designing experiments.
Asking a series of clear and focused questions
- The collaboration started at an advanced point of the project, when Senka and Jan understood what they wanted.
- At each step, they continued along the most logical, practical path. They were clear about why each new inquiry was worth pursuing.
- Although they “looked to the left and right” of the path, they didn’t try every possible analysis.
Working with mutual respect
- Senka and Jan’s data were well formatted, with sufficient metadata, and were ready for Janko’s pipeline.
- They respected each other’s time by turning tasks around quickly, and by having focused meetings only when necessary.
- They respected each other’s expertise, trusting each other to make decisions and engaging in constructive discussion.
Our results showed outstanding reproducibility of both community formation and evolution to steady state. Both SynCom and NatCom grew to community sizes representative of natural forest soils. Repeated community transfers into fresh soil systems led to rapid community stabilization across all ten biological replicates of SynCom, with similar trends observed in NatCom. This indicates strong deterministic forces shaping microbiome assembly and provides a base for more mechanistic examination of the nature of these forces. It also establishes these communities as reliable testbeds for controlled experimentation, for instance to probe the impact of stressors on soil communities.
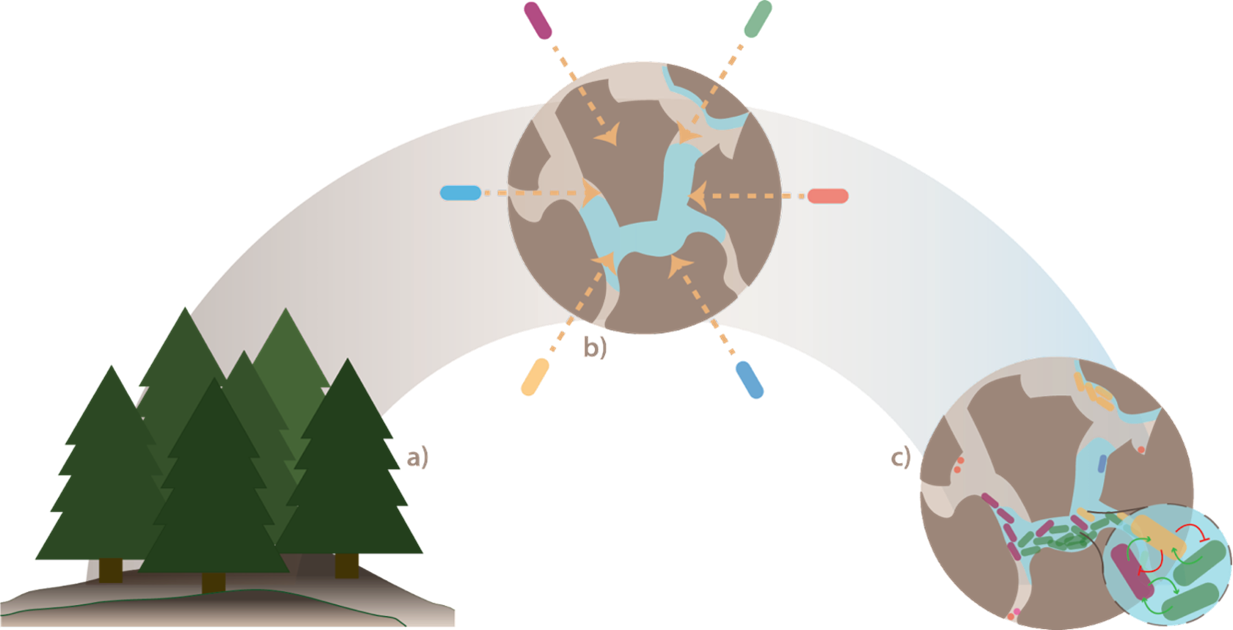
However, we did not want to stop here. In typical synthetic community design, one important question is frequently left unanswered: are the final synthetic communities still representative of the natural microbiomes? To this end, we leveraged Microbe Atlas (8) (www.microbeatlas.org), a global database developed by the von Mering group. The resource contains millions of environmental sequencing samples, spanning both major and exotic microbial habitats at a planetary scale. To ensure cross-study comparability, these community samples are mapped against a reference catalogue of curated marker genes, resulting in global abundance distributions of more than one hundred thousand species. Accompanied by rich, standardized metadata information, this allows users to both dive deep into particular environments and conditions or explore global trends. Scope, diversity, and comparability make it a unique playground for new science.
This comparison revealed that all SynComs and NatComs retained a global soil signature in the experiment. Interestingly, while the forest communities that our SynComs and NatComs were derived from clustered within typical forest/tundra samples, incubation pushed the communities closer towards agricultural soils. We suspect that the culturing process and nutrient concentration led to a rise of species typically associated with agricultural soil. Using the potential of Microbe Atlas, we found that most soil types are strongly distinguished by pH, which may therefore be an important driver for the community shift and an interesting factor for steering assembly (Figure 2). In the future, this collaborative approach of coupling synthetic community experiments with global microbiome comparisons may allow us to: identify key species, improve our synthetic communities, provide a wider context for locally observed phenomena, and broaden our scope to encompass different soil ecosystem types.
To conclude, our path to study the character of soil community assembly led us to i) design model communities which can be repurposed depending on the research question, ii) establish a reliable soil culturing system and, importantly, iii) find a way to relate synthetic microcosms back to natural communities. Throughout, the support of NCCR Microbiomes was crucial, as it brought two groups and two sets of expertise together. Despite never having met in person prior to article submission due to the pandemic, the collaboration was smooth and enjoyable, mainly due to the clear initial research question, as well as a pragmatic and goal-oriented mindset of everyone involved. We think many research questions could benefit from this synergy of experimental and computational approaches and encourage scientists to look out for such opportunities.
We thank Prof. Jan Roelof van der Meer and NCCR Microbiomes management team – Eavan Dorcey, Kendra Brown and Robin Tecon – for helpful feedback on this text.
Senka Causevic is a PhD Student at the University of Lausanne.
Janko Tackmann is a Senior Assistant at the University of Zurich.
References:
- Čaušević S, Tackmann J, Sentchilo V, von Mering C, van der Meer JR. Reproducible Propagation of Species-Rich Soil Bacterial Communities Suggests Robust Underlying Deterministic Principles of Community Formation. Msystems. 2022;7(2):e00160-22.
- Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nature Reviews Microbiology. 2017;15(10):579-90.
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology. 2006;4(2):102-12.
- Delgado-Baquerizo M, Oliverio AM, Brewer TE, Benavent-González A, Eldridge DJ, Bardgett RD, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359(6373):320-5.
- Egidi E, Delgado-Baquerizo M, Plett JM, Wang J, Eldridge DJ, Bardgett RD, et al. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat Commun. 2019;10(1):2369.
- Zolla G, Badri DV, Bakker MG, Manter DK, Vivanco JM. Soil microbiomes vary in their ability to confer drought tolerance to Arabidopsis. Applied Soil Ecology. 2013;68:1-9.
- Mishra GK. Microbes in Heavy Metal Remediation: A Review on Current Trends and Patents. Recent Pat Biotechnol. 2017;11(3):188-96.
- Rodrigues JFM, Tackmann J, Rot G, Schmidt TSB, Malfertheiner L, Danaila M, Gaio D, Näpflin N, von Mering C. The Microbe Atlas Project: a global survey of microbial communities (www.microbeatlas.org). Manuscript in preparation. 2023.